gjam
latest release gjam 2.5.2 on 2022-02-15
Generalized Joint Attribute Modeling (GJAM) in R
Ecological attributes include species abundances, traits, and individual condition (e.g., growth or infection status), to name a few. They are multivariate data, but not all of one type. They can be combinations of presence-absence, ordinal, continuous, discrete, composition, or zero-inflated. gjam provides inference on sensitivity to input variables, correlations between responses, model selection, prediction of responses, inverse prediction of predictors, and community classification by response to predictors.
gjam was motivated by species distribution and abundance data, but can provide an attractive alternative to traditional methods wherever observations are multivariate and combine multiple scales and mixtures of continuous and discrete data.
Importantly, analysis is done on the observation scale. That is, coefficients and covariances are interpreted on the same scale as the data. This contrasts with standard generalized linear models (GLMs), where coefficients and covariances are difficult to interpret and cannot be compared across responses that are modeled on different scales and with nonlinear link functions.
gjam accommodates massive zeros in multivariate data by avoiding the standard mixtures used in zero-inflated GLMs. Instead, gjam relies on censoring.
gjam exploits censoring to combine multiple data types in a single model, including mixtures of continuous and discrete data. For example, the microbial community (composition data) might be tracked together with host condition (continuous, categorical, binary, ordinal, …).
Model:
Clark, J.S., D. Nemergut, B. Seyednasrollah, P. Turner, and S. Zhang. 2017. Generalized joint attribute modeling for biodiversity analysis: Median-zero, multivariate, multifarious data. Ecological Monographs, 87, 34–56. Clark2017EcolMonogr, clarksupplement.
Presents the motivation and model; summarizes computation in gjam. The Supplement file provides additional detail on algorithms.
Taylor-Rodríguez, D., Kaufeld, K., Schliep, E. M., Clark, J. S., Gelfand, A. E. 2017. Joint species distribution modeling: Dimension reduction using Dirichlet processes. Bayesian Analysis, doi: 10.1214/16-BA1031. http://projecteuclid.org/euclid.ba/1478073617 bayesanaly2016
Many applications require large numbers of response variables. Microbiome studies bring the additional complication of composition data. And most observed values can still be zero. This paper describes the Dirichlet process prior implemented in gjam that finds a low-dimensional representation for the covariance between responses.
Dynamic species interactions:
Clark, J. S., C. L. Scher, and M. Swift. 2020. The emergent interactions that govern biodiversity change. Proceedings of the National Academy of Sciences, 202003852, https://doi.org/10.1073/pnas.2003852117. clarkPNAS2003852117.full
Quantifying species interactions requires dynamic data and the integration of environmental effects. This paper defines and estimates environment-species interactions (ESI) with full uncertainty.
Collins, C.G., Elmendorf, S.C., Smith, J.G., Shoemaker, L., Szojka, M., Swift, M., and K. N. Suding. 2022. Global change re-structures alpine plant communities through interacting abiotic and biotic effects Ecology Letters, 25, 1813-1826. https://doi.org/10.1111/ele.14060
Density-dependent shifts in competitive interactions drove long-term changes in abundance of species-groups under global change while counteracting environmental drivers limited the growth response of the dominant species through density-independent mechanisms. Furthermore, competitive interactions shifted with the environment, primarily with nitrogen and drove non-linear abundance responses across environmental gradients.
Microbiome applications:
Wang, Z., D. L. Juarez, J.-F. Pan, S. K. Blinebry, J. Gronniger, J. S. Clark, Z. I. Johnson, and D. E. Hunt. 2019. Microbial communities across nearshore to offshore coastal transects are shaped by both distance from shore and seasonality. Environmental Microbiology, 21, 3862-3872.
16S rRNA gene libraries reveal distinct nearshore, continental shelf, and offshore oceanic communities. Water temperature and distance from shore both most influence community composition. However, at the phylotype level, the distribution of some taxa is linked to temperature, others to distance from shore and some by both.
Bachelot B., Uriarte M., Muscarella R., Forero-Montana J., Thompson J., McGuire K., Zimmerman J.K., Swenson N.G. and J.S. Clark. 2018. Associations among arbuscular mycorrhizal fungi and seedlings are predicted to change with tree successional status. Ecology 99: 607-620.
Seedlings of early‐successional tree species may not rely as much as mid‐ and late‐successional species on AM fungi, and AM fungi may accelerate forest succession.
Trait analysis:
Whitworth, A., Beirne, C., Flatt, E., Froese, G., Nuñez, C. and Forsyth, A. 2021. Recovery of dung beetle biodiversity and traits in a regenerating rainforest: a case study from Costa Rica’s Osa Peninsula. Insect Conserv Divers, 14: 439-454. https://doi.org/10.1111/icad.12470.
Dung beetles are used here to assess tropical biodiversity patterns and recovery in human-modified forests. We conducted a comprehensive dung beetle survey (coprophagous and necrophagous communities) within five habitat types, across a land-use gradient, in the ecologically biodiverse Osa Peninsula, in Costa Rica’s south Pacific. Under favourable conditions, secondary rainforest recovered similar levels of species richness, and key traits of old-growth forest dung beetle communities. However, at the community-level, dung beetle abundance, richness, biomass, and diversity varied between habitat types of different anthropogenic disturbance and land-use. Regenerating secondary growth and plantation forests showed community compositions similar to old growth forests, while open and fragmented habitats had degraded and impoverished levels of dung beetle biodiversity.
Clark, J.S. 2016. Why species tell us more about traits than traits tell us about species: Predictive models. Ecology,97, 1979–1993, ecology2016, ecology2016_AppendixS1
The joint distribution of ecological attributes (‘traits’) can be modeled together with species, separately, or predicted from the joint distribution of species. This paper describes the model and computation implemented in gjam.
Niche shifts across life history stages:
Qiu, T., S. Shubhi, C. W. Woodall, and J.S. Clark. 2021. Niche shifts from trees to fecundity to recruitment that determine species response to climate change. Frontiers in Ecology and Evolution 9, 863. https://www.frontiersin.org/article/10.3389/fevo.2021.719141, DOI10.3389/fevo.2021.719141.
Shifts in niche requirements across adult to fecundity to tree regeneration are evaluated for North American tree species to show the strong temperature dependence in fecundity relative to more important local habitat variables for adults and tree recruitment.
Fisheries:
Roberts, S., A. M. Boustany, P. Halpin, and J.S. Clark. 2022. Jointly modeling marine species to inform the effects of environmental change on an ecological community in the Northwest Atlantic. Scientific Reports, in press.
Prediction:
Nunez, C.L., J. R. Poulsen, L.J.T. White, V. Medjibe, and J. S. Clark. 2022. Distinct community-wide responses to forecasted climate change in Afrotropical forests. Frontiers in Ecology and Evolution, in press.
Using country-wide tree species inventories, remotely sensed climate data, and future climate predictions from Gabon we predict a 3 – 8% decrease in Afrotropical forest species richness by the end of the century. This contrasts with a 30 – 50% loss of plant diversity predicted for equivalent warming in the Neotropics, showing that community forecasts are not generalizable across regions, and more representative studies are needed in understudied diverse biomes.
Vignette with R code and applications: gjam vignette
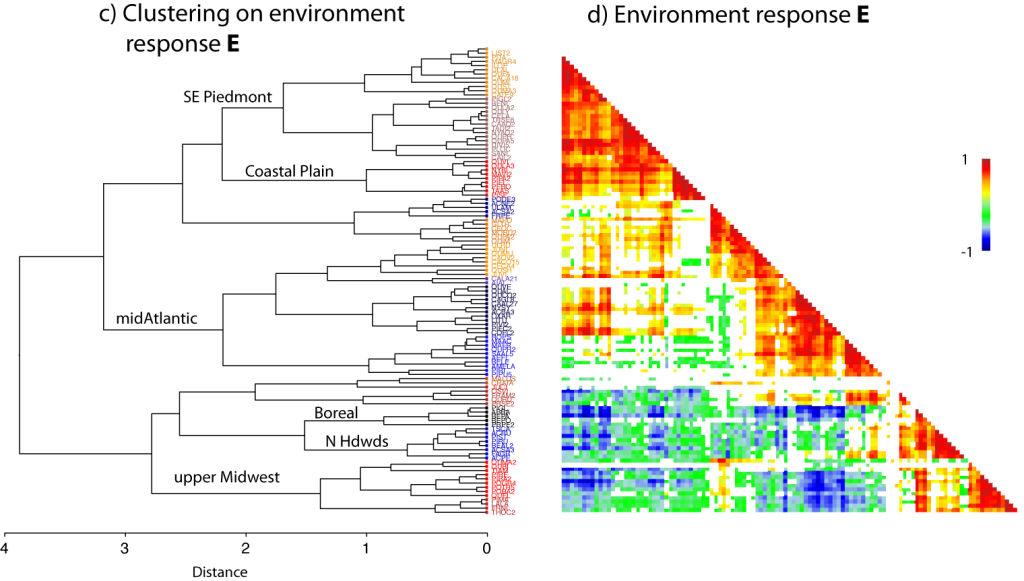
Below are cluster plots of the correlation matrix for a presence-absence model (a), continuous abundance model (b), and the response to environmental variables (d). The cluster analysis in (c) is based on distances in (d).
Main contributors:
Jim Clark wrote the GJAM model, the R and C++ code, and the GJAM package.
Alan Gelfand and Daniel Taylor-Rodrigues wrote the Dirichlet process model and algorithms for dimension reduction.
Daniel Taylor-Rodrigues implemented the Dirichlet process in R and C++ in GJAM.
Bene Bachelot, Chase Nuñes, and Brad Tomasek provided extensive testing and feedback through all stages of development.
Many others: Students in the course Bayesian Inference Environm Models (BIO/ENV 665) at Duke University and members of the Multivariate Modeling working group of the SAMSI Ecology program contributed many ideas, recommendations, and feedback.
Installation in R or RStudio:
> install.packages('gjam')
> library('gjam')
Documentation:
> help('gjam')
> browseVignettes('gjam')
Publications using GJAM
Whitworth, A., Beirne, C., Flatt, E., Froese, G., Nuñez, C. and Forsyth, A. (2021), Recovery of dung beetle biodiversity and traits in a regenerating rainforest: a case study from Costa Rica’s Osa Peninsula. Insect Conservation and Diversity. https://doi.org/10.1111/icad.12470
Clark, J. S., C. L. Scher, and M. Swift. 2020. The emergent interactions that govern biodiversity change. Proceedings of the National Academy of Sciences, .
Wang, Z., D. L. Juarez, J.-F. Pan,2, S. K. Blinebry, J. Gronniger, J. S. Clark, Z. I. Johnson, and D. E. Hunt. 2019. Microbial communities across nearshore to offshore coastal transects are shaped by both distance from shore and seasonality. Environmental Microbiology, 21(10):3862-3872.doi: 10.1111/1462-2920.14734.
Bachelot B., Uriarte M., Muscarella R., Forero-Montana J., Thompson J., McGuire K., Zimmerman J.K., Swenson N.G. and J.S. Clark. 2018. Associations among arbuscular mycorrhizal fungi and seedlings are predicted to change with tree successional status. Ecology, 99: 607-620. https://doi.org/10.1002/ecy.2122
Clark, J.S. 2016. Why species tell us more about traits than traits tell us about species: Predictive models. Ecology,97, 1979–1993,
Clark, J.S., D. Nemergut, B. Seyednasrollah, P. Turner, and S. Zhang. 2017. Generalized joint attribute modeling for biodiversity analysis: Median-zero, multivariate, multifarious data. Ecological Monographs, 87, 34–56.
Taylor-Rodríguez, D., Kaufeld, K., Schliep, E. M., Clark, J. S., Gelfand, A. E. 2017. Joint species distribution modeling: Dimension reduction using Dirichlet processes. Bayesian Analysis, doi: 10.1214/16-BA1031. http://projecteuclid.org/euclid.ba/1478073617