mastif
mast inference and forecasting (mastif)
Clark, J.S., C. Nunez, and B. Tomasek. 2019. Foodwebs based on unreliable foundations: spatio-temporal masting merged with consumer movement, storage, and diet. Ecological Monographs, in press. Appendix
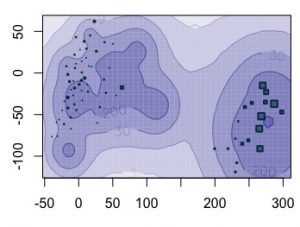
mastif is used to estimate fecundity and seed dispersal based on mapped tree and seed trap data. mastif operates on inventory plot data or simulated data. Inventory data for seeds of the genus Pinus are shown below for random plots and years. In these maps, large green symbols indicate seed traps that accumulated large numbers of seeds. Large brown symbols indicate large trees.
The simulator in mastif produces data sets with known parameter values and latent variables. Analysis provides insight on identifiability of seed production and the variables that control it.
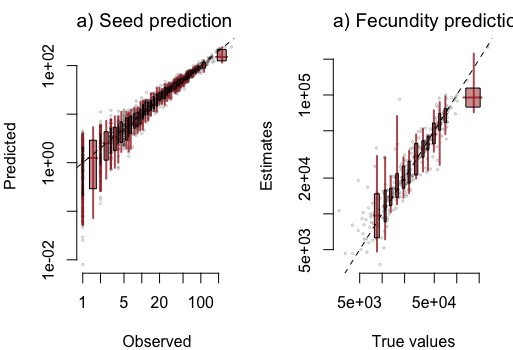
mastSim.Prediction validates the model and shows how well parameters and latent states are identified for specific data sets.
Maps of predicted seed rain and uncertainty from the fitted model. Predicted mean seed rain at left and predictive coefficient of variation at right. In-sample prediction includes mapped stands fitted with the model. Out-of-sample prediction is available for mapped stands that are not fitted to the data.

Year effects show ‘masting’, the tendency for individuals to synchronize reproduction. Year effects can be fitted by species and by species and region.
mastifuses Gibbs sampling with direct sampling, Metropolis, and Hamiltonian updating. It is implemented in R and C++ with Rcpp and the RcppArmadillo library for fast, efficient linear algebra operations.
Vignette with R code and applications
Package on CRAN now
Documentation in R:
> help('mastif')
> browseVignettes('mastif')
